Electrophoresis: Difference between revisions
imported>Sarah Richardson |
mNo edit summary |
||
| (30 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
{{subpages}} | {{subpages}} | ||
'''Electrophoresis''' | '''Electrophoresis''' involves the migration of particles or molecules (in particular proteins, [[DNA]], and [[RNA]]) through an [[electric field]] that separates them exclusively on the basis of their size or [[molecular weight]]. The direction the molecule moves depends on its charge while the rate of migration is affected by the size, shape, density of the gel and the strength of the applied current. | ||
Electrophoresis is a very simple process and relatively quick with a high resolution. In addition electrophoresis is an extremely useful method to estimate the purity of a sample. The technique is also very sensitive to slight variations in molecular weight, size, and even shape of nucleic acids and proteins <ref name=Harrison>Harrison, R. G., Todd, P., Rudge S. R., Petrides, D. P. (2003). Bioseparations Science and Engineering. New York, NY: Oxford University Press.</ref>. Electrophoresis can also be useful when it doesn’t affect the molecule’s structure or denature the protein <ref name = Harrison /ref>. | Electrophoresis is a very simple process and relatively quick with a high [[resolution]]. In addition electrophoresis is an extremely useful method to estimate the purity of a sample. The technique is also very sensitive to slight variations in molecular weight, size, and even shape of nucleic acids and proteins <ref name=Harrison>Harrison, R. G., Todd, P., Rudge S. R., Petrides, D. P. (2003). Bioseparations Science and Engineering. New York, NY: Oxford University Press.</ref>. Electrophoresis can also be useful when it doesn’t affect the molecule’s structure or denature the protein <ref name = Harrison /ref>. | ||
| Line 9: | Line 9: | ||
== The Process == | == The Process == | ||
The velocity of the particles are related to the electric field potential by the following equation: | {{Image|Picture_7.png|right|250px|Electrophoresis Apparatus}} | ||
As shown in the diagram, the nucleic acids or proteins are loaded into the wells or depressions at one end on the eletrophoretic medium (also known as a ‘’gel’’). The apparatus also has two electrodes on either side of the eletrophoretic medium. The [[anode]] is positively charged while the [[cathode]] is negatively charged. When a power source connects the two electrodes the charged particles begin to migrate towards the oppositely charged electrode due to the [[electric potential field]] within the media <ref name=Harrison />. | |||
The velocity of the particles are related to the [[electric field potential]] by the following equation: | |||
:<math>uμ = \frac{v}{E}\ </math> | :<math>uμ = \frac{v}{E}\ </math> | ||
Where E is the electric field potential that provides the driving force on the particle. μ is the electrophoretic | Where E is the [[electric field potential]] that provides the driving force on the particle. μ is the [[electrophoretic mobilit]]y and v is the velocity of the particle <ref name=Harrison />. For proteins, the equation can also be written as | ||
:<math>uμ = \frac{v}{E}\ = \frac{Z}{f}\ </math> | :<math>uμ = \frac{v}{E}\ = \frac{Z}{f}\ </math> | ||
Where Z is the protein’s net charge and f is a frictional coefficient related to the protein’s shape <ref name=Nelson>Nelson, D. L., Cox, M. M. (2008). Lehninger Principles of Biochemistry (5th ed.). New York, NY: W.H. Freeman and Company. | Where Z is the protein’s [[net charge]] and f is a [[frictional coefficient]] related to the protein’s shape <ref name=Nelson>Nelson, D. L., Cox, M. M. (2008). Lehninger Principles of Biochemistry (5th ed.). New York, NY: W.H. Freeman and Company. | ||
</ref>. | </ref>. | ||
| Line 26: | Line 30: | ||
==Generation of Heat in Electrophoresis Instrumentation== | ==Generation of Heat in Electrophoresis Instrumentation== | ||
Due to the electric field in electrophoresis, the equipment generates a large amount of heat that needs to be dissipated for maximum efficiency. Since the gel’s viscosity and density changes with an increasing temperature, it is important to remove as much heat as possible from the apparatus otherwise the gel will melt. As a solution, increasing the surface area to volume ratio of the gel usually helps to dissipate the heat. For instance, capillary electrophoresis efficiently removes heat because of its high surface-area to volume ratio. Similar to native electrophoresis, this commonly used method maintains a constant electric field at a stable pH where the separation depends upon mobility <ref name=Harrison />. | Due to the [[electric field]] in electrophoresis, the equipment generates a large amount of heat that needs to be dissipated for maximum [[efficiency]]. Since the gel’s [[viscosity]] and [[density]] changes with an increasing temperature, it is important to remove as much heat as possible from the apparatus otherwise the gel will melt. As a solution, increasing the surface area to volume ratio of the gel usually helps to dissipate the heat. For instance, [[capillary electrophoresis]] efficiently removes heat because of its high surface-area to volume ratio. Similar to native electrophoresis, this commonly used method maintains a constant electric field at a stable [[pH]] where the separation depends upon mobility <ref name=Harrison />. | ||
== Electrophoretic Mediums == | == Electrophoretic Mediums == | ||
One of the major factors affecting the separation of nucleic acids or proteins is the density of the gel. Polyacrylamide Gels are the most commonly used gels for electrophoresis and are mainly used to separate smaller proteins or fragments of nucleic acids (with molecular weights as low as 2000). Agarose gels on the other hand are used to separate much larger molecules (above a molecular weight of 106) <ref name=RickwoodP >Rickwood, D. & B.D. Hames (Eds.). (1982). Gel electrophoresis of proteins: a practical approach. Oxford: IRL Press Limited.</ref>. As the percentage of agarose or polyacrylamide increases in the solution, the gel becomes denser. As a result, the nucleic acids or proteins move slower since the pore size has decreased. The resolution or distance between the fragments (or bands) relative to their width, therefore, depends on the density of the gel <ref name=RickwoodP />. The density of the gel chosen is usually one that adequately separates all the components of interest. | One of the major factors affecting the separation of nucleic acids or proteins is the [[density]] of the gel. [[Polyacrylamide Gels]] are the most commonly used gels for electrophoresis and are mainly used to separate smaller proteins or fragments of [[nucleic acids]] (with molecular weights as low as 2000). [[Agarose gels]] on the other hand are used to separate much larger molecules (above a molecular weight of 106) <ref name=RickwoodP >Rickwood, D. & B.D. Hames (Eds.). (1982). Gel electrophoresis of proteins: a practical approach. Oxford: IRL Press Limited.</ref>. As the percentage of agarose or polyacrylamide increases in the solution, the gel becomes denser. As a result, the nucleic acids or proteins move slower since the pore size has decreased. The resolution or distance between the fragments (or bands) relative to their width, therefore, depends on the density of the gel <ref name=RickwoodP />. The density of the gel chosen is usually one that adequately separates all the components of interest. | ||
== Detection Techniques == | == Detection Techniques == | ||
There are several detection techniques used to visualize bands. The most commonly used technique involves chemically staining proteins with coomassie blue or silver. Another method covalently bonds proteins to fluorescent molecules before electrophoresis. | There are several detection techniques used to visualize bands. The most commonly used technique involves chemically staining proteins with [[coomassie blue]] or silver. Another method [[covalently bonds]] proteins to [[fluorescent]] molecules before electrophoresis. | ||
Nucleic acids are commonly visualized by illuminating the bands under a UV light after binding them to ethidium bromide <ref name=RickwoodP />. Other methods include: chemical staining, fluorescence, radioactivity, immunoelectrophoretic techniques and on-column/end-column detection <ref name=Harrison />. | Nucleic acids are commonly visualized by illuminating the bands under a [[UV light]] after binding them to [[ethidium bromide]] <ref name=RickwoodP />. Other methods include: [[chemical staining]], [[fluorescence]], [[radioactivity]], [[immunoelectrophoretic techniques]] and [[on-column/end-column detection]] <ref name=Harrison />. | ||
| Line 41: | Line 45: | ||
== Gel Electrophoresis of DNA and RNA == | == Gel Electrophoresis of DNA and RNA == | ||
Negatively charged phosphate ions on nucleic acids (DNA and RNA) give them an overall negative net charge. As a result, the negatively charged nucleic acids will migrate towards the positive anode in the gel thereby separating the molecules. The bands can then be visualized under UV light if ethidium bromide was added to the nucleic acid before beginning electrophoresis <ref name=Chen />. Usually a photograph of the gel is taken in order to approximate the quantity of the fragment. | Negatively charged [[phosphate ions]] on [[nucleic acids]] ([[DNA]] and [[RNA]]) give them an overall negative net charge. As a result, the negatively charged nucleic acids will migrate towards the positive anode in the gel thereby separating the molecules. The bands can then be visualized under [[UV light]] if [[ethidium bromide]] was added to the nucleic acid before beginning electrophoresis <ref name=Chen />. Usually a photograph of the gel is taken in order to approximate the quantity of the fragment. | ||
There are two ways to determine an unknown fragment’s molecular weight in a gel. | There are two ways to determine an unknown fragment’s [[molecular weight]] in a gel. | ||
Either a rough estimate can be made by comparing the unknown fragment’s length to the size standards in the left column of the photograph (as shown in figure) or by using a standard curve. In most cases, the distance moved by a nucleic acid in a gel is inversely related to its molecular weight. After completing electrophoresis on a sample of nucleic acids with known molecular weights a standard curve can be created by plotting the log of the migration distances vs. the log of the molecular weights. Since the resulting curve is linear the unknown fragment’s length can then be determined <ref name=RickwoodNA>Rickwood, D. & B.D. Hames (Eds.). (1981). Gel electrophoresis of nucleic acids: a practical approach. Oxford: IRL Press Limited.</ref>. | Either a rough estimate can be made by comparing the unknown fragment’s length to the size standards in the left column of the photograph (as shown in figure) or by using a [[standard curve]]. In most cases, the distance moved by a nucleic acid in a gel is inversely related to its molecular weight. After completing electrophoresis on a sample of nucleic acids with known [[molecular weights]] a [[standard curve]] can be created by plotting the log of the migration distances vs. the log of the [[molecular weights]]. Since the resulting curve is linear the unknown fragment’s length can then be determined <ref name=RickwoodNA>Rickwood, D. & B.D. Hames (Eds.). (1981). Gel electrophoresis of nucleic acids: a practical approach. Oxford: IRL Press Limited.</ref>. | ||
== Gel Electrophoresis of Proteins == | == Gel Electrophoresis of Proteins == | ||
| Line 51: | Line 55: | ||
== Native Electrophoresis == | == Native Electrophoresis == | ||
Molecules are separated in their native form (not denatured) at a constant pH with a constant electric field. Separation depends upon mobility. This process uses a pH around 8 or 9 where most proteins are negatively charged so that they move towards the anode. The bands can then be visualized by staining the proteins with coomassie blue or silver or by adding fluorescent protein labels <ref name=Harrison />. | Molecules are separated in their native form (not denatured) at a constant [[pH]] with a constant [[electric field]]. Separation depends upon mobility. This process uses a [[pH]] around 8 or 9 where most proteins are negatively charged so that they move towards the [[anode]]. The bands can then be visualized by staining the proteins with [[coomassie blue]] or silver or by adding [[fluorescent]] protein labels <ref name=Harrison />. | ||
== Denaturing Gel electrophoresis == | == Denaturing Gel electrophoresis == | ||
Denaturing gel electrophoresis is done with a constant electric field at a constant pH. Since it is necessary for proteins to have a similar charge to mass ratio during electrophoresis, sodium dodecyl sulfate (SDS) is added to the solution. SDS binds to the positively charged residues of the proteins and gives them an overall net negative charge. Otherwise the proteins may not migrate at the same rate or in the same direction on the gel due to their different intrinsic charges. As a result, the negatively charged proteins bound to SDS will migrate towards the positive side of the electric field in the gel <ref name=Nelson />. However, as a consequence SDS also denatures the proteins so that mobility in the gel depends only on molecular weight. Therefore, gel electrophoresis separates proteins exclusively on the basis of molecular weight, without any interference from charge. | {{Image|Picture_12.png|right|250px|}} | ||
Denaturing gel electrophoresis is done with a constant [[electric field]] at a constant [[pH]]. Since it is necessary for proteins to have a similar charge to mass ratio during electrophoresis, [[sodium dodecyl sulfate]] (SDS) is added to the solution. SDS binds to the positively charged residues of the proteins and gives them an overall net negative charge. Otherwise the proteins may not migrate at the same rate or in the same direction on the gel due to their different intrinsic charges. As a result, the negatively charged proteins bound to SDS will migrate towards the positive side of the electric field in the gel <ref name=Nelson />. However, as a consequence SDS also denatures the proteins so that mobility in the gel depends only on [[molecular weight]]. Therefore, gel electrophoresis separates proteins exclusively on the basis of [[molecular weight]], without any interference from charge. | |||
Similar to native electrophoresis, the protein bands are stained with coomassie blue or a fluorescent protein label is added. Analysis of the gel provides a rough estimate of the concentration of proteins in the solution or the number of subunits in the protein <ref name=Nelson />. Once again, the approximate molecular weight of an unknown protein can be determined by comparing it against proteins of known molecular weight. | Similar to native electrophoresis, the protein bands are stained with [[coomassie blue]] or a [[fluorescent]] protein label is added. Analysis of the gel provides a rough estimate of the [[concentration]] of proteins in the solution or the number of subunits in the protein <ref name=Nelson />. Once again, the approximate molecular weight of an unknown protein can be determined by comparing it against proteins of known molecular weight. | ||
=== | === Disadvantages/Advantages === | ||
Separating proteins by gel electrophoresis is not an efficient method to purify large amounts of proteins since there are much better methods. The process is also very difficult to scale. However, gel electrophoresis is a great way to monitor the purity of a protein after each purification process <ref name=Nelson />. | Separating proteins by gel electrophoresis is not an efficient method to purify large amounts of proteins since there are much better methods. The process is also very difficult to [[scale]]. However, gel electrophoresis is a great way to monitor the purity of a protein after each purification process <ref name=Nelson />. | ||
== Isoelectric | == Isoelectric Focusing == | ||
Isoelectric focusing separates proteins based on their isoelectric points (pI) by establishing a pH gradient within the gel. Ampholytes are distributed across the gel to form the pH gradient <ref name=Nelson />. Once the proteins are added at one end of the gel they will begin to migrate towards the positively charged anode due to the constant electric field. The proteins will continue moving until their isolectric point (pI) matches the pH, at which point they will have zero net charge. This technique is mainly used to determine the isoelectric point of an unknown protein and to separate proteins with the same molecular weight but with different pI values. | {{Image|Picture_8.png|right|250px|Isoelectric Focusing}} | ||
Isoelectric focusing separates proteins based on their [[isoelectric points]] (pI) by establishing a [[pH]] gradient within the gel. [[Ampholytes]] are distributed across the gel to form the [[pH]] gradient <ref name=Nelson />. Once the proteins are added at one end of the gel they will begin to migrate towards the positively charged [[anode]] due to the constant [[electric field]]. The proteins will continue moving until their [[isolectric point]] (pI) matches the [[pH]], at which point they will have zero net charge. This technique is mainly used to determine the [[isoelectric point]] of an unknown protein and to separate proteins with the same [[molecular weight]] but with different pI values. | |||
== Two-Dimensional Electrophoresis for Proteins == | == Two-Dimensional Electrophoresis for Proteins == | ||
Two-dimensional electrophoresis encompasses both isoelectric focusing and gel electrophoresis. After separating the proteins by isoelectric focusing, the first gel is turned sideways and placed in the second gel where the proteins begin to migrate according to their size. | {{Image|Picture_10.png|right|250px|2D Electrophoresis for Proteins}} | ||
Using two-dimensional separation one can quickly estimate both the molecular weight and the isoelectric point of a protein. More importantly, this technique can also be used to distribute proteins throughout a gel with identical molecular weights but different pI values or vice versa <ref name=Nelson />. This method is also extremely useful for solutions with a large number of proteins or a mixture containing similar proteins. Although, the process cannot be used for proteins that are extremely acidic or basic, where their pI is outside the pH range <ref name=RickwoodP />. However, the main disadvantage in using this method is comigration of proteins (or the aggregation of proteins) that arise due to interactions with each other <ref name=RickwoodP />. | Two-dimensional electrophoresis encompasses both [[isoelectric focusing]] and gel [[electrophoresis]]. After separating the proteins by [[isoelectric focusing]], the first gel is turned sideways and placed in the second gel where the proteins begin to migrate according to their size. | ||
Using two-dimensional separation one can quickly estimate both the [[molecular weight]] and the [[isoelectric point]] of a protein. More importantly, this technique can also be used to distribute proteins throughout a gel with identical [[molecular weights]] but different pI values or vice versa <ref name=Nelson />. This method is also extremely useful for solutions with a large number of proteins or a mixture containing similar proteins. Although, the process cannot be used for proteins that are extremely [[acidic]] or [[basic]], where their pI is outside the pH range <ref name=RickwoodP />. However, the main disadvantage in using this method is [[comigration]] of proteins (or the [[aggregation]] of proteins) that arise due to interactions with each other <ref name=RickwoodP />. | |||
== Two-Dimensional Electrophoresis for DNA and RNA == | == Two-Dimensional Electrophoresis for DNA and RNA == | ||
The following table summarizes the different methods used to separate DNA or RNA fragments by two-dimensional electrophoresis <ref name=RickwoodNA />. | The following table summarizes the different methods used to separate [[DNA]] or [[RNA]] fragments by two-dimensional electrophoresis <ref name=RickwoodNA />. | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 79: | Line 86: | ||
| DNA | | DNA | ||
| Conventional Gels | | Conventional Gels | ||
| Uses an Agarose-acrylamide gel | | Uses an [[Agarose-acrylamide gel]] | ||
| Polyacrylamide gel at 50°C | | [[Polyacrylamide gel]] at 50°C | ||
|- | |- | ||
| DNA | | DNA | ||
| Denaturing Gradient Gels | | Denaturing Gradient Gels | ||
| Agarose gel | | [[Agarose gel]] | ||
| Polyacrylamide gel with a urea gradient | | [[Polyacrylamide gel]] with a [[urea gradient]] | ||
|- | |- | ||
| RNA | | RNA | ||
| Urea Shift | | [[Urea Shift]] | ||
| Constant gel conc., Neutral pH range, Low urea conc. | | Constant gel conc., Neutral [[pH]] range, Low urea conc. | ||
| Constant gel conc., Neutral pH range, High urea conc. | | Constant gel conc., Neutral [[pH]] range, High urea conc. | ||
|- | |- | ||
| RNA | | RNA | ||
| Concentration Shift | | Concentration Shift | ||
| Constant urea conc., Neutral pH range, Low gel conc. | | Constant urea conc., Neutral [[pH]] range, Low gel conc. | ||
| Constant urea conc., Neutral pH range, Double the gel conc. | | Constant urea conc., Neutral [[pH]] range, Double the gel conc. | ||
|- | |- | ||
| RNA | | RNA | ||
| Urea, pH and Concentration Shift | | Urea, [[pH]] and Concentration Shift | ||
| Low gel conc., Acidic pH, High urea conc. | | Low gel conc., [[Acidic]] [[pH]], High urea conc. | ||
| Double the gel conc., Neutral pH, Zero urea conc. | | Double the gel conc., Neutral [[pH]], Zero urea conc. | ||
|} | |} | ||
== Blotting Techniques == | == Blotting Techniques == | ||
The three blotting techniques that involve transferring proteins or nucleic acids from a gel to nitrocellulose paper are Southern blotting, Northern blotting and Western blotting. The following table summarizes each method. Blotting is mainly useful for identify a specific molecule on a gel by probing with a ligand. | The three blotting techniques that involve transferring proteins or nucleic acids from a gel to [[nitrocellulose paper]] are [[Southern blotting]], [[Northern blotting]] and [[Western blotting]]. The following table summarizes each method. Blotting is mainly useful for identify a specific molecule on a gel by probing with a [[ligand]]. | ||
{| class="wikitable" | |||
|- | |||
! Technique | |||
! [[Molecules]] | |||
! [[Probe]] | |||
! Detection | |||
|- | |||
| [[Southern Blotting]] | |||
| [[DNA]] | |||
| [[Radiolabeled]] or [[nonradiolabeled]] complementary [[RNA]] or [[DNA]] | |||
| [[Autoradiography fluorescence]], [[chemiluminescence]], or [[colorimetry]] | |||
|- | |||
| [[Northern Blotting]] | |||
| [[RNA]] | |||
| [[Radiolabeled]] or [[nonradiolabeled]] complementary [[RNA]] or [[DNA]] | |||
| [[Autoradiography fluorescence]], [[chemiluminescence]], or [[colorimetry]] | |||
|- | |||
| [[Western Blotting]] | |||
| [[Proteins]] | |||
| Whole [[antisera]] or [[antibodies]] | |||
| [[Autoradiography fluorescence]], [[chemiluminescence]], or [[colorimetry]] | |||
|} | |||
In Southern blotting, single stranded DNA fragments that have been distributed throughout a gel by electrophoresis are transferred to nitrocellulose paper. This is done by essentially laying the nitrocellulose paper on top of the gel and allowing capillary action to pull the alkaline solution and DNA upward through the gel <ref name=Campbell>Campbell, N. A., Reece, J. B., Urry, L. A., Cain, M. L., Wasserman, S. A., Minorshky, P. V., et al. (2008). Biology (8th ed.). San Francisco, CA: Benjamin Cummings.</ref>. Once all the DNA fragments have been transferred, a solution containing a radioactively labeled probe (usually) is added to the nitrocellulose blot <ref name=Campbell />. The single stranded DNA or RNA probe then binds to its complementary DNA fragment in the matrix. Bands containing DNA that base-paired with the probe can then be easily detected <ref name=Campbell />. The same procedure is used in Northern and Western blotting, except the probe and detection method vary. | In [[Southern blotting]], single stranded [[DNA]] fragments that have been distributed throughout a gel by [[electrophoresis]] are transferred to [[nitrocellulose paper]]. This is done by essentially laying the [[nitrocellulose paper]] on top of the gel and allowing [[capillary action]] to pull the [[alkaline]] solution and [[DNA]] upward through the gel <ref name=Campbell>Campbell, N. A., Reece, J. B., Urry, L. A., Cain, M. L., Wasserman, S. A., Minorshky, P. V., et al. (2008). Biology (8th ed.). San Francisco, CA: Benjamin Cummings.</ref>. Once all the [[DNA]] fragments have been transferred, a solution containing a [[radioactively]] labeled probe (usually) is added to the [[nitrocellulose]] blot <ref name=Campbell />. The single stranded [[DNA]] or [[RNA]] probe then binds to its complementary [[DNA]] fragment in the matrix. Bands containing [[DNA]] that base-paired with the probe can then be easily detected <ref name=Campbell />. The same procedure is used in Northern and [[Western blotting]], except the probe and detection method vary. | ||
==References== | ==References== | ||
<references/> | <references/>[[Category:Suggestion Bot Tag]] | ||
Revision as of 06:01, 11 August 2024
For the course duration, the article is closed to outside editing. Of course you can always leave comments on the discussion page. The anticipated date of course completion is 21 December 2010. One month after that date at the latest, this notice shall be removed. Besides, many other Citizendium articles welcome your collaboration! Note to course participants: Looking forward to some insightful and useful articles from your collaborations. |
Electrophoresis involves the migration of particles or molecules (in particular proteins, DNA, and RNA) through an electric field that separates them exclusively on the basis of their size or molecular weight. The direction the molecule moves depends on its charge while the rate of migration is affected by the size, shape, density of the gel and the strength of the applied current.
Electrophoresis is a very simple process and relatively quick with a high resolution. In addition electrophoresis is an extremely useful method to estimate the purity of a sample. The technique is also very sensitive to slight variations in molecular weight, size, and even shape of nucleic acids and proteins [1]. Electrophoresis can also be useful when it doesn’t affect the molecule’s structure or denature the protein Cite error: Invalid <ref> tag; invalid names, e.g. too many
The Process
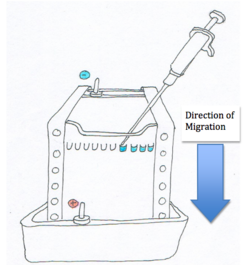
As shown in the diagram, the nucleic acids or proteins are loaded into the wells or depressions at one end on the eletrophoretic medium (also known as a ‘’gel’’). The apparatus also has two electrodes on either side of the eletrophoretic medium. The anode is positively charged while the cathode is negatively charged. When a power source connects the two electrodes the charged particles begin to migrate towards the oppositely charged electrode due to the electric potential field within the media [1].
The velocity of the particles are related to the electric field potential by the following equation:
- Failed to parse (syntax error): {\displaystyle uμ = \frac{v}{E}\ }
Where E is the electric field potential that provides the driving force on the particle. μ is the electrophoretic mobility and v is the velocity of the particle [1]. For proteins, the equation can also be written as
- Failed to parse (syntax error): {\displaystyle uμ = \frac{v}{E}\ = \frac{Z}{f}\ }
Where Z is the protein’s net charge and f is a frictional coefficient related to the protein’s shape [2].
Smaller molecules move faster in the gel than larger molecules and therefore they end up closer to the positive anode. Molecules that are about the same size move at the same rate through the electrophoretic medium. The figure to the right shows the molecules in ‘’bands’’. The column on the far left contains bands with known molecular weights. This is useful to determine the molecular weight of an unknown particle [2].
Generation of Heat in Electrophoresis Instrumentation
Due to the electric field in electrophoresis, the equipment generates a large amount of heat that needs to be dissipated for maximum efficiency. Since the gel’s viscosity and density changes with an increasing temperature, it is important to remove as much heat as possible from the apparatus otherwise the gel will melt. As a solution, increasing the surface area to volume ratio of the gel usually helps to dissipate the heat. For instance, capillary electrophoresis efficiently removes heat because of its high surface-area to volume ratio. Similar to native electrophoresis, this commonly used method maintains a constant electric field at a stable pH where the separation depends upon mobility [1].
Electrophoretic Mediums
One of the major factors affecting the separation of nucleic acids or proteins is the density of the gel. Polyacrylamide Gels are the most commonly used gels for electrophoresis and are mainly used to separate smaller proteins or fragments of nucleic acids (with molecular weights as low as 2000). Agarose gels on the other hand are used to separate much larger molecules (above a molecular weight of 106) [3]. As the percentage of agarose or polyacrylamide increases in the solution, the gel becomes denser. As a result, the nucleic acids or proteins move slower since the pore size has decreased. The resolution or distance between the fragments (or bands) relative to their width, therefore, depends on the density of the gel [3]. The density of the gel chosen is usually one that adequately separates all the components of interest.
Detection Techniques
There are several detection techniques used to visualize bands. The most commonly used technique involves chemically staining proteins with coomassie blue or silver. Another method covalently bonds proteins to fluorescent molecules before electrophoresis.
Nucleic acids are commonly visualized by illuminating the bands under a UV light after binding them to ethidium bromide [3]. Other methods include: chemical staining, fluorescence, radioactivity, immunoelectrophoretic techniques and on-column/end-column detection [1].
Staining is an easy way to make a rough estimate of the quantity, purity and molecular weight of a compound. In general, the intensity of the staining is proportional to the concentration of the compound [4]. Generally, darker bands contain a larger quantity.
Gel Electrophoresis of DNA and RNA
Negatively charged phosphate ions on nucleic acids (DNA and RNA) give them an overall negative net charge. As a result, the negatively charged nucleic acids will migrate towards the positive anode in the gel thereby separating the molecules. The bands can then be visualized under UV light if ethidium bromide was added to the nucleic acid before beginning electrophoresis [4]. Usually a photograph of the gel is taken in order to approximate the quantity of the fragment.
There are two ways to determine an unknown fragment’s molecular weight in a gel.
Either a rough estimate can be made by comparing the unknown fragment’s length to the size standards in the left column of the photograph (as shown in figure) or by using a standard curve. In most cases, the distance moved by a nucleic acid in a gel is inversely related to its molecular weight. After completing electrophoresis on a sample of nucleic acids with known molecular weights a standard curve can be created by plotting the log of the migration distances vs. the log of the molecular weights. Since the resulting curve is linear the unknown fragment’s length can then be determined [5].
Gel Electrophoresis of Proteins
Gel electrophoresis can also be used to separate charged proteins in an electric field. The two principle methods to separate proteins are native gel electrophoresis and denaturing gel electrophoresis [1].
Native Electrophoresis
Molecules are separated in their native form (not denatured) at a constant pH with a constant electric field. Separation depends upon mobility. This process uses a pH around 8 or 9 where most proteins are negatively charged so that they move towards the anode. The bands can then be visualized by staining the proteins with coomassie blue or silver or by adding fluorescent protein labels [1].
Denaturing Gel electrophoresis
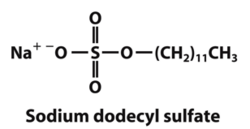
Denaturing gel electrophoresis is done with a constant electric field at a constant pH. Since it is necessary for proteins to have a similar charge to mass ratio during electrophoresis, sodium dodecyl sulfate (SDS) is added to the solution. SDS binds to the positively charged residues of the proteins and gives them an overall net negative charge. Otherwise the proteins may not migrate at the same rate or in the same direction on the gel due to their different intrinsic charges. As a result, the negatively charged proteins bound to SDS will migrate towards the positive side of the electric field in the gel [2]. However, as a consequence SDS also denatures the proteins so that mobility in the gel depends only on molecular weight. Therefore, gel electrophoresis separates proteins exclusively on the basis of molecular weight, without any interference from charge.
Similar to native electrophoresis, the protein bands are stained with coomassie blue or a fluorescent protein label is added. Analysis of the gel provides a rough estimate of the concentration of proteins in the solution or the number of subunits in the protein [2]. Once again, the approximate molecular weight of an unknown protein can be determined by comparing it against proteins of known molecular weight.
Disadvantages/Advantages
Separating proteins by gel electrophoresis is not an efficient method to purify large amounts of proteins since there are much better methods. The process is also very difficult to scale. However, gel electrophoresis is a great way to monitor the purity of a protein after each purification process [2].
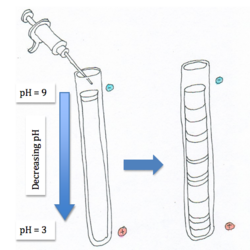
Isoelectric Focusing
Isoelectric focusing separates proteins based on their isoelectric points (pI) by establishing a pH gradient within the gel. Ampholytes are distributed across the gel to form the pH gradient [2]. Once the proteins are added at one end of the gel they will begin to migrate towards the positively charged anode due to the constant electric field. The proteins will continue moving until their isolectric point (pI) matches the pH, at which point they will have zero net charge. This technique is mainly used to determine the isoelectric point of an unknown protein and to separate proteins with the same molecular weight but with different pI values.
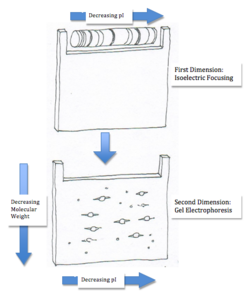
Two-Dimensional Electrophoresis for Proteins
Two-dimensional electrophoresis encompasses both isoelectric focusing and gel electrophoresis. After separating the proteins by isoelectric focusing, the first gel is turned sideways and placed in the second gel where the proteins begin to migrate according to their size. Using two-dimensional separation one can quickly estimate both the molecular weight and the isoelectric point of a protein. More importantly, this technique can also be used to distribute proteins throughout a gel with identical molecular weights but different pI values or vice versa [2]. This method is also extremely useful for solutions with a large number of proteins or a mixture containing similar proteins. Although, the process cannot be used for proteins that are extremely acidic or basic, where their pI is outside the pH range [3]. However, the main disadvantage in using this method is comigration of proteins (or the aggregation of proteins) that arise due to interactions with each other [3].
Two-Dimensional Electrophoresis for DNA and RNA
The following table summarizes the different methods used to separate DNA or RNA fragments by two-dimensional electrophoresis [5].
| Method | First Dimension | Second Dimension | |
|---|---|---|---|
| DNA | Conventional Gels | Uses an Agarose-acrylamide gel | Polyacrylamide gel at 50°C |
| DNA | Denaturing Gradient Gels | Agarose gel | Polyacrylamide gel with a urea gradient |
| RNA | Urea Shift | Constant gel conc., Neutral pH range, Low urea conc. | Constant gel conc., Neutral pH range, High urea conc. |
| RNA | Concentration Shift | Constant urea conc., Neutral pH range, Low gel conc. | Constant urea conc., Neutral pH range, Double the gel conc. |
| RNA | Urea, pH and Concentration Shift | Low gel conc., Acidic pH, High urea conc. | Double the gel conc., Neutral pH, Zero urea conc. |
Blotting Techniques
The three blotting techniques that involve transferring proteins or nucleic acids from a gel to nitrocellulose paper are Southern blotting, Northern blotting and Western blotting. The following table summarizes each method. Blotting is mainly useful for identify a specific molecule on a gel by probing with a ligand.
| Technique | Molecules | Probe | Detection |
|---|---|---|---|
| Southern Blotting | DNA | Radiolabeled or nonradiolabeled complementary RNA or DNA | Autoradiography fluorescence, chemiluminescence, or colorimetry |
| Northern Blotting | RNA | Radiolabeled or nonradiolabeled complementary RNA or DNA | Autoradiography fluorescence, chemiluminescence, or colorimetry |
| Western Blotting | Proteins | Whole antisera or antibodies | Autoradiography fluorescence, chemiluminescence, or colorimetry |
In Southern blotting, single stranded DNA fragments that have been distributed throughout a gel by electrophoresis are transferred to nitrocellulose paper. This is done by essentially laying the nitrocellulose paper on top of the gel and allowing capillary action to pull the alkaline solution and DNA upward through the gel [6]. Once all the DNA fragments have been transferred, a solution containing a radioactively labeled probe (usually) is added to the nitrocellulose blot [6]. The single stranded DNA or RNA probe then binds to its complementary DNA fragment in the matrix. Bands containing DNA that base-paired with the probe can then be easily detected [6]. The same procedure is used in Northern and Western blotting, except the probe and detection method vary.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Harrison, R. G., Todd, P., Rudge S. R., Petrides, D. P. (2003). Bioseparations Science and Engineering. New York, NY: Oxford University Press.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Nelson, D. L., Cox, M. M. (2008). Lehninger Principles of Biochemistry (5th ed.). New York, NY: W.H. Freeman and Company.
- ↑ 3.0 3.1 3.2 3.3 3.4 Rickwood, D. & B.D. Hames (Eds.). (1982). Gel electrophoresis of proteins: a practical approach. Oxford: IRL Press Limited.
- ↑ 4.0 4.1 Chen, K., Glase, J. (2009-2010). Chapter 13 – DNA Technology: From Recombination to Genomic Sequencing and Forensic Analysis. In Chen, K. & Hester, L. L. (Eds.), Investigative Biology a laboratory text (pp. 245-288). Plymouth, MI: Hayden McNeil.
- ↑ 5.0 5.1 Rickwood, D. & B.D. Hames (Eds.). (1981). Gel electrophoresis of nucleic acids: a practical approach. Oxford: IRL Press Limited.
- ↑ 6.0 6.1 6.2 Campbell, N. A., Reece, J. B., Urry, L. A., Cain, M. L., Wasserman, S. A., Minorshky, P. V., et al. (2008). Biology (8th ed.). San Francisco, CA: Benjamin Cummings.