Parainfluenza virus: Difference between revisions
imported>Shabana Ansari No edit summary |
imported>Shabana Ansari |
||
| Line 11: | Line 11: | ||
==Description and significance== | ==Description and significance== | ||
Upper and lower respiratory tract diseases in humans are mostly caused by Human parainfluenze viruses and as a group are called “parainfluenza.” Human parainfluenza viruses (HPIVs) are the second to the respiratory syntial virus (RSV) as a common cause of upper and lower respiratory tract infections especially in infants and young children. <ref> http://en.wikipedia.org/wiki/Parainfluenza_virus </ref> It is less common among adults. | |||
Paramyxoviruses | Paramyxoviruses belong to Paramyxoviridae family of Mononegavirales order which are negative sense single stranded RNA (-ssRNA) viruses responsible for many diseases in humans and animals. Negative sense RNA first of all makes positive sense RNA through copying their genomes by RNA polymerase. The positive sense RNA molecule is translated into proteins acting as viral mRNA. Thus more –ssRNA are produced when the resultant proteins directs the synthesis of new virions such as capsid proteins and RNA repilcases. <ref>http://en.wikipedia.org/wiki/Negative-sense_virus</ref> | ||
==Genome structure== | ==Genome structure== | ||
Revision as of 22:21, 17 April 2009
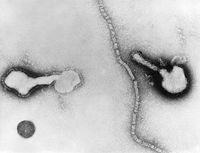
| Human Parainfluenza Virus | ||
|---|---|---|
 | ||
| Virus classification | ||
|
Description and significance
Upper and lower respiratory tract diseases in humans are mostly caused by Human parainfluenze viruses and as a group are called “parainfluenza.” Human parainfluenza viruses (HPIVs) are the second to the respiratory syntial virus (RSV) as a common cause of upper and lower respiratory tract infections especially in infants and young children. [1] It is less common among adults. Paramyxoviruses belong to Paramyxoviridae family of Mononegavirales order which are negative sense single stranded RNA (-ssRNA) viruses responsible for many diseases in humans and animals. Negative sense RNA first of all makes positive sense RNA through copying their genomes by RNA polymerase. The positive sense RNA molecule is translated into proteins acting as viral mRNA. Thus more –ssRNA are produced when the resultant proteins directs the synthesis of new virions such as capsid proteins and RNA repilcases. [2]
Genome structure
Cell structure and metabolism
Ecology
Pathology
Application to Biotechnology
Current Research
References
Be sure to replace "Needs" in "Needs Workgroup" below with a workgroup name. See the "Workgroups" link on the left for a list of workgroups.