Parainfluenza virus
| Human Parainfluenza Virus | ||
|---|---|---|
 | ||
| Virus classification | ||
|
Description and significance
Upper and lower respiratory tract diseases in humans are mostly caused by Human Parainfluenze Viruses and as a group are called “Parainfluenza.” [1] Human Parainfluenza Viruses (HPIVs) are the second to the Respiratory Synctial Virus (RSV) as a common cause of upper and lower respiratory tract infections especially in infants and young children. [2] It is less common among adults. HPIVs are Paramyxoviruses. Paramyxoviruses belong to Paramyxoviridae family of Mononegavirales order which are negative sense single stranded RNA (-ssRNA) viruses responsible for many diseases in humans and animals. [3] Negative sense RNA first of all makes positive sense RNA through copying their genomes by RNA polymerase. The positive sense RNA molecule is translated into proteins acting as viral mRNA. Thus more –ssRNA are produced when the resultant proteins directs the synthesis of new virions such as capsid proteins and RNA repilcases. [4]
Genome structure
HPIVs are negative sense single stranded RNAs (-ssRNAs).
Cell structure and metabolism
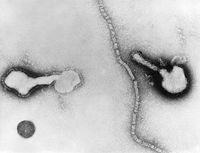
The Human Parainfluenza Viruses are consisting of a nulceocapsid and an envelope with an average diameter of 120-300 nm. The nucleocapsid is a tubelike structure with helical symmetry and 12-18 nm in width. It contains a –ssRNA molecule with 5-8*106 molecular weight, the phoshphopritein P, the major nucleoprotein NP and the RNA polymerase L protein. The L protein is necessary for viral transcription, P proteins takes part in RNA synthesis and NP helps to maintain the genome structure. The envelope is a double layered membrane. The outer layer is covered with spikes which consist of lipoproteins and glycoproteins whereas the inner layer contains non glycosylated matrix protein M. The spikes’s glycoproteins contain hemagglutinin neuraminidase HN and the cell fusion protein F. [5] Viruses acquire the energy from host cells to grow and reproduce. Hemagglutinin in the HPIV’s outer layer spikes attaches the virus to the host cell’s neuraminic acid receptor and F protein facilitates the fusion of virus into the host cell membrane. Since –ssRNA cannot serve as a messenger, L protein, a transcriptase, transcribes the positive sense RNA from the viral genome (-ssRNA). The positive sense RNA serves as mRNA and P protein facilitates the protein synthesis by ribosomes and more –ssRNA strands are produced which are enveloped in double layered membrane. It is stated, “P protein directs the viral protein synthesis and are copied into negative-sense RNA strands which are integrated in the new virions. For envelopment, the virus-specific glycoproteins accumulate in the cell membrane. Assembly is completed by budding of the nucleocapsid through the cell membrane studded with glycoproteins.” [6]
Ecology
HPIVs are pathogens and cause number of diseases since they belong to paramyxoviridae family and all members of this family cause significant diseases. Kelly J. Henrickson says about the paramyxovridae family, “Infact this virus family is one of the most costly in terms of disease burden and economic impact to our planet.” (p : 243) [7] HPIV is strongly affected by environmental conditions such as temperature, pH, humidity and the composition of the medium in which it is stored/grown. As temperature increases from 37°C, its survival chances decreases and it is inactivated within 15 minutes as the temperature approaches to 50°C. It is stable at less temperature such as 4°C and even when it is frozen at -70°C. Freezing might kill the virus or may harm its infectivity but even if very small amount of infectivity is survived, it is enough for the virus to recover. It can be recovered even after 26 years of being frozen. Reagents like bovine serum albumin, skim milk, dimethyl sulfoxide can help the virus in surviving for a long time if they are added into the virus’s culture before freezing. The optimal pH for the Virus’s stability is physiologic pH 7.0 to 8.0 while it loses its stability at ph 3.0 to 3.4, under low humidity and also with virus desiccation. [8]
Pathology
Application to Biotechnology
Current Research
References
- ↑ http://www.nlm.nih.gov/medlineplus/ency/article/001370.htm#Causes,%20incidence,%20and%20risk%20factors
- ↑ http://en.wikipedia.org/wiki/Parainfluenza_virus
- ↑ http://en.wikipedia.org/wiki/Paramyxovirus
- ↑ http://en.wikipedia.org/wiki/Negative-sense_virus
- ↑ http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mmed.section.3128
- ↑ http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mmed.section.3128
- ↑ Kelly J. Henrickson, Parainfluenza Viruses, Clinical Microbiology Reviews, April 2003, Vol 16. No 2, p. 243
- ↑ Kelly J. Henrickson, Parainfluenza Viruses, Clinical Microbiology Reviews, April 2003, Vol 16. No 2, p. 245
Be sure to replace "Needs" in "Needs Workgroup" below with a workgroup name. See the "Workgroups" link on the left for a list of workgroups.