Clostridium difficile
Articles that lack this notice, including many Eduzendium ones, welcome your collaboration! |
| Clostridium difficile | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Clostridium difficile |
Clostridium difficile is a bacteria that is a "common inhabitant of the colon flora in human infants and sometimes in adults. It produces a toxin that causes pseudomembranous enterocolitis in patients receiving antibiotic therapy."[1]
Classification
Higher order taxa[2]
Prokaryote; Firmicutes; Clostridia; Clostridiales; Clostridiaceae
Species[2]
Clostridium difficile
Description and significance
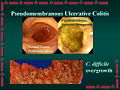
Clostridium difficile is a species of bacteria that is the leading cause of infectious diarhhea among patients in hospitals worldwide. C. difficile causes pseudomembranous enterocolitis, a plaque-forming infection of the colon that leads to heavy diarrhea and may be life-threatening. C. difficile is present at low levels in the gut flora of about 3% of adults. These people however show no symptoms and do not need to be treated. The infection occurs when a person is treated with antibiotics targeted against other bacteria. The disease is for the most part nosocomial. Patients who are hospitalized come in contact and are often inoculated with the bacteria. When the patient is treated with antibiotics, especially those with a broad range of activity, the normal gut flora is disrupted, and C. difficile, with its multi-drug resistance, experiences overgrowth. The bacteria releases large quantities of enterotoxins (toxin A) and cytotoxins (toxin B), causing pseudomembranous enterocolitis. To treat infection, antibiotic intake must cease and anticlostridial antibiotics (vancomycin, amoxicillin, ciprofloxacin, metronidazole, doxycycline, gentamicin, clindamycin) must be ingested.[2]
In 1935, Hall and O’Toole first isolated the bacteria from the stools of newborns and described it. They named it Bacillus difficilis because it was hard to isolate and grew very slowly in culture.[3]
C. difficile is an important pathogen that is currently increasing in its prevalence world-wide. A complete genome sequence would enable geneticists to come up with a more direct and efficient treatment against the pathogen. The genetic material encodes for antimicrobial resistance, production of toxins (virulence), host interaction (adaptations for survival and growth within the gut environment), and the production of surface structures. The understanding of how these genes interact with their environment will be useful in developing therapies against C. difficile associated diseases.[4]
Genome Structure
Sebaihia et al (2006) determined the complete genomic sequence of C. difficile strain 630, a highly virulent and multidrug-resistant strain. It was found that the genome consists of a circular chromosome of 4,290,252 bp and a plasmid, pCD630, of 7,881 bp. The chromosome encodes 3,776 predicted coding sequences (CDSs), with resistance, virulence, and host interaction genes, while the plasmid carries only 11 CDSs, none of which has any obvious function. C. difficile has a highly mobile genome, with 11% of the genome consisting of mobile genetic elements, mostly in the form of conjugative transposons. Conjugative transposons are mobile genetic elements that are capable of integrating into and excising from the host genome and transferring themselves, and are responsible for the evolutionary acquisition by C. difficile of genes involved in resistance, virulence, and host interactions. Some of the mobile elements are prophage sequences. Host interaction genes involve genes that code for metabolic capability adaptations for survival and growth within the gut environment.[4]
The genome carries several copies of a very interesting genetic element, the IStron. The IStron is a hybrid between an intron and an insertion sequence (IS), and can therefore insert itself into a DNA sequence, and later be excised from the primary mRNA transcript by cellular machinery. For example, a copy inserted in the tcdA gene (that encodes the enterotoxin A) renders the gene nonfunctional until the IStron is excised. Truncated copies of the IStron (containing only the IS sequence) were found in intergenic regions. Possibly, these truncated variants may be the answer to make C. difficile unable to produce functional toxins. [4]
Cell structure and metabolism
Clostridia are Gram-positive, anaerobic, spore-forming, rod-shaped, motile bacteria. As spores, the bacteria look like drumsticks, with a bulge located at one end. C. difficile produces an S layer (polysaccharide capsule) that contributes to its pathogenicity. C. difficile has flagellae that contribute to its motility.[2]
C. difficile is an obligate fermenter, and exhibits optimal growth in blood agar at human body temperatures in the absence of oxygen. The metabolism of C. difficile is in large part adapted to life in the intestinal tract.[2] It produces enzymes that degrade nutrients abundant in the intestine. Carbohydrates are the preferred nutrient source, and C. difficile has the ability to metabolize a wide range of carbohydrates. The bacteria also have a relatively unique ability to utilize ethanolamine, an abundant phospholipid provided by the host’s dietary intake, as a carbon and nitrogen source.[4]
C. difficile has the enzyme that catalyzes the decarboxylation of p-hydroxyphenylacetate (a tyrosine degradation product) to p-crysol, a compound that stunts bacterial growth. C. difficile produces and tolerates high concentrations of p-crysol, giving it a competitive advantage over the normal bacterial flora in the intestine.[4]
Ecology
C. difficile is found throughout nature, especially in soil. It exhibits optimal growth in blood agar at human body temperatures in the absence of oxygen. It can remain dormant in hospitals in the form of spores until patient inoculation. Following treatment of antibiotics, C. difficile overgrows in the intestinal tract, causing pseudomembranous enterocolitis, and consequently diarrhea.[2]
Pathology
C. difficile is transmitted from person to person in a fecal-oral fashion in the form of vegetative cells and heat-resistant spores. Most vegetative cells are killed in the stomach, but spores pass through the stomach unaffected because of their acid resistance. Spores germinate in the small bowel upon exposure to bile acids. C. difficile multiplies in the colon and overgrows in the absence of competitors (following antibiotic treatment). Mucosa of the gut allows C. difficile adherence to the colonic epithelium.[5]
Different pathogenic strains of C. difficile produce different toxins, but the most common ones are enterotoxin (toxin A) and cytotoxin (toxin B). The two toxins lead to "the production of tumour necrosis factor-alpha and pro-inflammatory interleukins, increased vascular permeability, neutrophil and monocyte recruitment, opening of epithelial cell junctions and epithelial cell apoptosis." Hydrolytic enzymes cause connective tissue to degrade, leading to colitis, pseudomembrane formation and watery diarrhea.[5]
Clostridium difficile infection (CDI) can range in severity from asymptomatic to life threatening. Death is more common among the aged (>65). Patient symptoms include severe diarrhea, exposure to antibiotics, foul stool odor, and abdominal pain.[5]
Research
There is an ongoing search for new therapeutic strategies to battle the ever-increasing pathogenicity of the C. difficile strains. Barreto et al (2008) have recently released a study on the effects of the polysaccharide fucoidin, an L-selectin blocker, on toxin-A-induced mouse enteritis. Enteritis is medical term that refers to the inflammation of the intestine, usually accompanies by diarrhea. Fucoidin or the placebo (saline) was injected into the mouse, and 5 minutes later, toxin A was locally released. To measure the effect of fucoidin, intestinal fluid volume/length and ileal loop weight/length ratios were calculated three hours later. It was found that fucoidin significantly (P < 0.05) reduced the toxin-A-induced increase in weight/length and volume/length ratios and reduced mucosal disruption.[6]
Gerber and Ackermann (2008) have published a report on another treatment option, OPT-80. OPT-80 (otherwise known as tiacumicin B, lipiarmycin or PAR-101) is “an antimicrobial with little or no systemic absorption after oral administration and narrow activity spectrum against Gram-positive aerobic and anaerobic bacteria.” OPT-80 is highly effective at very low concentrations, making it a very safe and efficient drug. More clinical trials are to be completed, but for now results look promising.[7]
The reason why C. difficile-associated disease (CDAD) is becoming such a widespread nosocomial disease is because the spores it forms are resistant to heat, chemicals, antibiotics, and radiation, making the hospital environment difficult to sanitize. The study of C. difficile germination from spores to vegetative cells will shed light on the strengths and weaknesses of the spores, and will thus lead to the development of methods for spore eradication. Sorg and Sonenshein (2008) studied the effects of the components of bile on the germination of C. difficile spores. They found that cholate derivatives and glycine act to induce germination in the duodenum. It was found that, interestingly, deoxycholate, a metabolite of cholate (produced by the normal intestinal flora) induces germination while preventing growth of vegetative cells. This implies a mechanism by which normal gut flora can mediate the levels of vegetative C. difficile cells in the gut.[8]
References
- ↑ Anonymous (2024), term (English). Medical Subject Headings. U.S. National Library of Medicine.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Clostridium difficile. 12 Apr. 2008. Wikipedia, the free encyclopedia. 13 Apr. 2008. <http://en.wikipedia.org/wiki/Clostridium_difficile>
- ↑ Hall I, O'Toole E (1935). "Intestinal flora in newborn infants with a description of a new pathogenic anaerobe, Bacillus difficilis". Am. J. Dis. Child 49: 390.
- ↑ 4.0 4.1 4.2 4.3 4.4 Sebaihia M et al (2006). "The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome". Na. Gen. 38: 779–86. PMID 16804543. [e]
- ↑ 5.0 5.1 5.2 C. difficile. 12 Apr. 2008. The Writing Lab and OWL at Purdue and Purdue University. 10 March 2006. <http://media.romanvenable.net/images/cDiffLarge.jpeg>
- ↑ Barreto AR et al (2008). "Fucoidin Prevents Clostridium difficile Toxin-A-Induced Ileal Enteritis in Mice". Dig. Dis. Sci. 53(4): 990-6. PMID 17805968. [e]
- ↑ Gerber M, Ackermann G (2008). "OPT-80, a macrocyclic antimicrobial agent for the treatment of Clostridium difficile infections: a review". Expert. Opin. Investig. Drugs 17(4): 547–53. PMID 18363518. [e]
- ↑ Sorg JA, Sonenshein AL (2008). "Bile salts and glycine as cogerminants for Clostridium difficile spores". J Bacteriol 190(7): 2505-12. PMID 18245298. [e]