Streptococcus mutans: Difference between revisions
imported>Young Okeke No edit summary |
imported>Young Okeke |
||
| Line 39: | Line 39: | ||
==Genome structure== | ==Genome structure== | ||
S. mutans has a single double stranded circular genome. The sequence of the 2,030,936 bp Streptococcus mutans strain UA159 Genome is completed and the results published in the October 29, 2002 issue of Proceedings National Academy of Sciences USA. . This feat was accomplished first sequencing 12,000 individual, shotgun-based, double stranded templates. Next, a directed custom synthetic primer-based approach was used for closure and quality improvement to complete the sequence of the genome to a high level of accuracy. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Revision as of 21:26, 15 April 2008
Articles that lack this notice, including many Eduzendium ones, welcome your collaboration! |
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Streptococcus mutans |
Description and significance
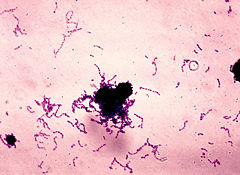
The genus Streptococcus is defined mainly in terms of the morphology of the cells. These bacteria are gram-positive spherical cells that appear mainly in chains due to cell division in the same plane. Incomplete separation of divided cells produces the chain of cells which vary in length. [2]
Streptococcus mutans was first described by JK Clark in 1924 after he isolated it from a carious legion, but it wasn’t until the 1960’s that a great deal of interest was generated when researchers began studying dental caries. [2]
The group of oral streptococci closely related to S. mutans is referred to as the “mutans group” or mutans streptococci. The actual taxonomic bacteria Streptococcus mutans is always written in italics while the group name mutans streptococci is not. [3] The mutans group consists of S. rattus, S. mutans, S. cricetus, S. maccacae, S. sobrinus, and S. downeii. [2] S. mutans and S. sobrinus comprise the majority of the group and are only found in humans. [2] They can be distinguished by laboratory tests, but it is not always practical due to costs and time. There is no selective media that would allow the detection and separation of the species and laboratory work is therefore done on the entire mutans streptococci group. Because of its greater prevalence, most of the isolates will in fact be S. mutans. [3]
The two selective media that are widely used for isolating caries-related streptococci are based on Mitis-Salivarius agar and TYC agar to which the antibiotic Bacitracin is added (TYSCB). This suppresses the growth of most species but allows S. mutans and S. sobrinus to grow. The inclusion of sucrose leads to the formation of glucans and a distinctive colony appearance that aids in identification. [3]
Describe the appearance, habitat, etc. of the organism, and why it is important enough to have its genome sequenced. Describe how and where it was isolated.
Include a picture or two (with sources) if you can find them.
Streptococcus mutans is involved in the initial formation of the biofilm involves components of the oral flora to form dental plaque on teeth. This flora is extensive and may reach a thickness of 300-500 cells on the surface of the teeth. [1]
S. mutans as well as other bacteria that compose the normal oral flora provide valuable services to the human host. They occupy available colonization sites which makes it more difficult for other microorganisms to establish themselves. The oral flora also contributes to host nutrition through the synthesis of vitamins, and they contribute to immunity by inducing low levels of circulating and secretory antibodies that have the potential to react with other pathogens. In addition, the oral flora exerts microbial antagonism against foreign species through the production of inhibitory substances. [1]
One of the harmful effects of the normal flora, especially S. mutans, is bacterial synergism between a member of the normal flora and a potential pathogen. During the bacterial synergism, a member of the normal flora facilitates the growth of other potential pathogens. S. mutans facilitates such a condition through the initiation of the biofilm to which several oral bacteria adhere to and rely upon. S. mutans readily colonizes tooth surfaces and forms a thin film on the tooth called the enamel pellicle. It contains a cell-bound protein, glycosyl transferase, that serves as an adhesion for attachment to the tooth and produces lactic acid which demineralizes tooth enamel. Furthermore, if oral streptococci such as S. mutans are introduced into wounds created by dental manipulation or treatment, they may adhere to heart valves and initiate subacute bacterial endocarditis. [1]
Genome structure
S. mutans has a single double stranded circular genome. The sequence of the 2,030,936 bp Streptococcus mutans strain UA159 Genome is completed and the results published in the October 29, 2002 issue of Proceedings National Academy of Sciences USA. . This feat was accomplished first sequencing 12,000 individual, shotgun-based, double stranded templates. Next, a directed custom synthetic primer-based approach was used for closure and quality improvement to complete the sequence of the genome to a high level of accuracy.
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
In utero the human fetal oral cavity is sterile, but the colonization of bacteria begins at birth. Handling and feeding of the infant after birth leads to the establishment of a stable normal flora in the oral cavity in about 48 hours. Streptococcus salivarius is the first dominant colonizer and may make up 98% of the total oral flora until the appearance of teeth occurs around 6 to 9 months of age. After the eruption of teeth, S. mutans and S. sanguis colonize the oral cavity and persist as long as teeth remain. [1]
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
Researchers are working to interfere with key genes and proteins necessary for the survival of S. mutans to remove the ability of the bacteria to thrive in acidic conditions. Past research has shown that this ability has several components including the bacterial membrane bound enzyme fatty acid byosynthase M (FabM), which when shut down makes S. mutans 10,000 times more vulnerable to acid damage. In addition, early work suggests that FabM helps to resist the human body’s defenses. As a result, FabM is a major target for the design of new drugs. [4]
Researchers are also working to identify and rank every one of the 2,000 known S. mutans genes that contribute to its fitness. [4]
References
[1] http://www.textbookofbacteriology.net/normalflora.html
[2] http://www.ncl.ac.uk/dental/oralbiol/oralenv/tutorials/streps.htm
[3] http://www.ebi.ac.uk/2can/genomes/bacteria/Streptococcus_mutans.html