Magnaporthe grisea
| Magnaporthe grisea
A conidium and conidiogenous cell of M. grisea | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Magnaporthe grisea |
Description and significance
Magnaporthe grisea, a plant-pathogenic fungus, is the causal agent of rice blast disease which is one of the greatest pathological threats to rice crops. It was thought to be localized only in developing nations, however for the past decade it has emerged as a severe problem in the United States, more recently in California. Yearly, rice that can feed an estimated 60 million people is destroyed by this disease. M.grisea is a well adapted fungus that can attack and penetrate its host plant with ease. More importantly, this fungus can also infect other important agricultural crops including cereals such as wheat, rye, pearl millet, and barley. The disease that is cause is then referred as blight disease or blast disease. Furthermore, M.grisea is thought of as a model organism in studies of fungal phytopathogenicity and host-parasite interactions[1].
Genome structure
Due to the destructive nature of this pathogen, sequencing the entire genome was necessary to understand the mechanisms of how the fungus causes disease. The complete genome of M.grisea was sequenced in 2005. M. grisea is haploid and its genome encodes of ~40 Mb that are contained in 7 chromosomes.[2][3] The genome consists of a varied group of secreted proteins and a range of G-protein coupled receptor (GPCR) genes. Furthermore, strong phylogenetic evidence of horizontal gene transfer (HGT) has been found between M.grisea (ascomycete fungi) and oomycetes ( a distant relative)[4].
Cell structure and metabolism
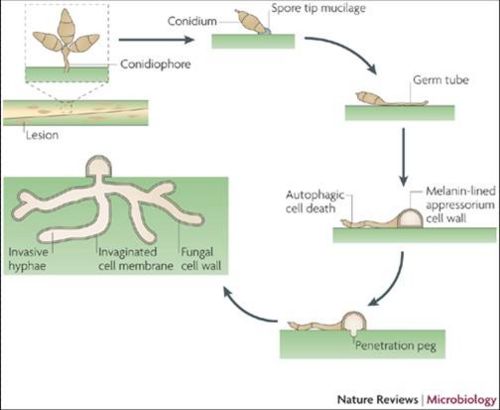
M.grisea is a filamentous fungus. The spore tips contains a spore tip mucilage (STM) adhesive that allows M.grisea to bind to its host[5]. The binding of the spore begins the infection progression. After host-binding, specialized infection cells, termed appresoria, provide mechanical force and thus penetrate the plant cuticle and gain entry into the internal tissues of the host.
M.grisea are infectious species and so obtain nutrients from their hosts. M.grisea is primarily haploid, and can reproduce sexually and asexually. The M.grisea infectious life cycle is asexual. The conidium is the infectious structure in this life cycle. After colonization of its host, lesions form. The lesion sites are locations of M.grisea sporulation. Nearby plants become infected from the conidia spores as well.
Ecology
M.grisea inhibits the host plant’s immune system, affecting the metabolism and cell signaling pathways. Adaptation of M.grisea is so efficient that even rice cultivars (bred to resist the disease) are infected. All above-ground parts of a host plant can be infected, but only if the stem, node, or panicle becomes infected will the grain set be lost. Rice is one of the most important staple foods worldwide and control of this disease is extremely important.
Disease Cycle
Pathology
High nitrogen content, host plant moisture, and 20°C of night temperatures favors the disease. There are many races of this fungal pathogen and their level of virulence varies [7]. Rice cultivars have been bred with the different resistance genes that have been identified, however the pathogen overcomes the resistance genes within a few years. It is hypothesized that resistance could be due to the high genetic variation of this pathogen[8].
Control Measures[9]
Burning/composting of diseased tissues - Diseased plants are burned or composted to prevent the spread of the disease to the next crop season.
Healthy Crop - For a healthy crop, healthy seeds are necessary and many precautions are to be taken. The seeds are collected from the field where M.grisea existence is unlikely due to hostile conditions for the pathogen. Fungicide and disinfectants are then applied if necessary for protection against a variety of pathogens.
Fertilizer management - Excess nitrogen fertilizer can facilitate the disease progression, whereas silica limits the development of the disease. The amount and type of fertilizer are to be discretionally applied.
Chemical control - The following fungicides are applied to combat the rice blast disease: benomyl, fthalide, edifenphos, iprobenfos, tricyclazole, isoprothiolane, probenazole, pyroquilon, felimzone(= meferimzone), diclocymet, carpropamid, fenoxanil and metominostrobin, and antibiotics such as blasticidin and kasugamycin.
Resistant cultivars - Based on studies and current knowledge of the different races of M.grisea, resistant cultivars have been bred throughout the world.
Current Research
"The Genome Sequence of the Rice Blast Fungus Magnaporthe grisea" [10]
-This study was conducted to sequence the whole genome of M.grisea. Presented in this paper are preliminary analyses of the genome. The genome was sequenced via a shotgun approach. The draft genome sequenced consists of 2,273 sequence contigs that are longer than 2kb. Total length of the genome is estimated to be ~40Mb. 11,109 genes are predicted to yield protein products and make up 48% of M.grisea genome. Evidently, phylogenitically none of the gene families of M.grisea shows recent lineage-specific expansion indicating that ancient gene duplication may have played a role followed by loss of the gene families in the other lineages. Dean and colleagues indicate that M.grisea has a considerable number of proteins that are diverse and novel in cell pathways. Furthermore, the generation of genetic variations is tremendous and could explain the efficiency of this fungal pathogen to infect resistant rice cultivars within a few generations.
"Cellular Differentiation and Host Invasion by the Rice Blast Fungus Magnaporthe grisea" [11]
-Current advances towards the understanding of the rice blast fungus are described in this article. The recent sequence of the genome provided much insight into the structure and number of genes that are required to cause the rice blast disease. Phylogenetic analysis provided evidence for multiple horizontal gene transfers. There is also a better understanding of the early stages of plant infections. Appresoria are the specialized structures that infect the plants. M.grisea can also infect the host plant roots. Within the roots, the fungus then can invade the vascular system and ultimately kill the plant. Furthermore, functional genomics studies have led to the discovery of ~200 genes that are essential for the rice blast disease. It is to be noted that though equipped with much knowledge of the biology of M.grisea, there are impediments to overcome.
"Roles for Rice Membrane Dynamics and Plasmodesmata during Biotrophic Invasion by the Blast Fungus" [12]
-This paper investigates the way M.grisea grows within the plant cells. The penetration peg of the appresorium pierces the host plants surface. The cytoplasm of the peg contains abundant actin microfilaments that extend into the lumen of the plants' epidermal cells. This extension then facilitates the movement of the appresorium nucleus and cytoplasmic contents. This primary growing hyphae then differentiates into an invasive hyphae (IH) that will progress into neighboring cells. Three distinct types of hyphae that play a role in biotrophic invasion have been identified in this paper: (1) filamentous primary hyphae in the first-invaded plant cell; (2) bulbous IH in first-invaded and in subsequently invaded plant cells; and (3) filamentous IH that initially grow in successively invaded cells. Additionally, supportive evidence is provided in regards to the cell-to-cell movement of the blast IH with the use of plasmodesmata. This is quite a significant discovery because mostly viruses, which are considerably smaller than M.grisea, are known to move this way.
References
- ↑ Broad Institute. "Magnaporthe Information." 24 June 2005. Accessed April 17, 2009.
- ↑ Talbot, N.J.; D.J. Ebbole & J.E. Hamer (1993), "Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast …", The Plant Cell Online 5 (11): 1575–1590. Retrieved on 2009-04-17
- ↑ Diaz-perez, S.V.; V.W. Crouch & M.J. Orbach (1996), "Construction and Characterization of a Magnaporthe grisea Bacterial Artificial Chromosome Library", Fungal Genetics and Biology 20 (4): 280–288. Retrieved on 2009-04-17
- ↑ Dean, R.A.; N.J. Talbot & D.J. Ebbole et al. (2005), "The genome sequence of the rice blast fungus Magnaporthe grisea", Nature 434 (7036): 980–986. Retrieved on 2009-04-17
- ↑ http://microbewiki.kenyon.edu/index.php/Magnaporthe
- ↑ > Wilson, R.A. and Talbot, N.J. (2009)"Under pressure: investigating the biology of plant infection by Magnaporthe oryzae", Nature 434 (7036)185-195. retrieved 2009-04-28
- ↑ Talbot, N.J. (2003), "On the trail of a cereal killer", Annu. Rev. Microbiol 57: 177–202. Retrieved on 2009-04-17
- ↑ Dean, R.A.; N.J. Talbot & D.J. Ebbole et al. (2005), "The genome sequence of the rice blast fungus Magnaporthe grisea", Nature 434 (7036): 980–986. Retrieved on 2009-04-17
- ↑ George N. Agrios "Plant Pathology" Edition: 5, illustrated, annotated Published by Academic Press, 2005 ISBN 0120445654, 9780120445653 922 pages, accessed 2009-04-17
- ↑ Dean, R.A.; N.J. Talbot & D.J. Ebbole et al. (2005), "The genome sequence of the rice blast fungus Magnaporthe grisea", Nature 434 (7036): 980–986. Retrieved on 2009-04-17
- ↑ Caracuel-rios, Z. & N.J. Talbot (2007), "Cellular differentiation and host invasion by the rice blast fungus Magnaporthe grisea", Current Opinion in Microbiology 10 (4): 339–345. Retrieved on 2009-04-17
- ↑ Kankanala, P.; K. Czymmek & B. Valent (2007), "Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus", The Plant Cell Online 19 (2): 706. Retrieved on 2009-04-17

