Myxococcus xanthus
| Myxococcus xanthus | ||||||
|---|---|---|---|---|---|---|
 | ||||||
| Scientific classification | ||||||
| ||||||
| Binomial name | ||||||
| Myxococcus xanthus |
Description and significance
Myxococcus xanthus is a social organism, which is self-organized, saprotrophic and predatory. M. xanthus is a rod shaped, Gram-negative bacteria, which uses a form of gliding for locomotion.[1] There's been recent discovery of two types of systems used for locomotion. The first system is type IV pilli, which is used as a type of hook. There's been recent discovery of two types of systems used for locomotion. The first system is type IV pilli, which is used as a type of hook. The second system is mucus secretion, which tends to form sites of focal adhesion. M. xanthus can also be defined as a social bacteria because of the cell to cell communication that it undergoes during predation and life cycle changes.[2] During starvation periods this bacteria has been discovered to use a form of chemotactic signaling in order to produce and regulate multi-cellular rippling during predation. Predation takes place in a multi-cellular synchronizing mechanism. The ability for this bacterium to communicate with others and be able to work together towards the targeted prey, makes it a good candidate for research. M. xanthus forms fruiting bodies. Within these fruiting bodies there are spores. These spores will germinate in order to return to a vegetative cycle as soon as conditions are favorable. M. xanthus is a predator to other bacteria, but harmless to humans. [3]
Genome structure
M. xanthus contains one of the largest prokaryotic genomes to be sequenced. The length of the genome is 9,139,763 nt, containing 7456 genes of which 7331 encode proteins, 79 are structural RNAs, and 43 are pseudo genes. The 9.14 Mb is contained on a single circular chromosome. It is also a bacterium that has been genetically followed in order to research genetic modification. [4]
Cell structure and metabolism
M. xanthus has a life cycle which includes predation, fruit bodies and swarming.[5] Fruit bodies usually form from the cause of starvation, and these bodies can then form into stress-resistant spores. These spores are in a dormant stage, where they will withstand long periods of starvation or any other environmental change such as: temperature and desiccation. Leucine is an important amino acid for the growth of this bacterium. The size of M. xanthus is about ten times bigger than E.coli.[6]
Ecology
M. xanthus is commonly found in high organic matter soil with preference to a range of pH from 5-8 and preferably damp. They thrive at higher levels of organic matter in soil. They have also been found in rocky surfaces. There have been studies done on the texture of agar affecting predation, whether in soft or hard agar no change occurs. This bacterium lives in a multi-cellular unit.[7]
Pathology
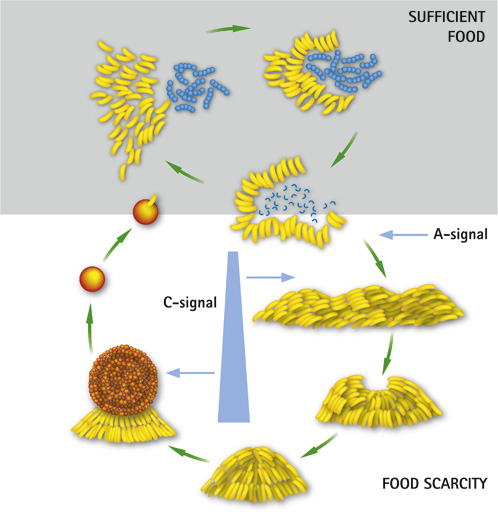
This bacteria is a predator of other bacteria, but non-pathogenic. The fact that M. xanthus moves by chemotaxis and usually moves towards its prey resulted in the word "predataxis".[8] M. xanthus is not a single unit when attacking a prey, rather there is a sense of cooperation. Predation of M. xanthus includes release of toxic and lytic substances. These substances can degrade or paralyze the targeted prey. Predation takes place by many cell to cell signaling happening, these signals are named A, B, C signals which are also used in the transformation of the life cycle. [9] The life cycle mainly relies on whether there is food available in the environment or not. if there is enough food available in the environment M.xanthus will predate and swarm in their environment. During periods where there is not food available, M.xanthus will form fruiting bodies, which will then morph into stress-resistant spores. These spores are the dormant part of the life cycle, until there is enough food available in the environment.[10]
Application to biotechnology
The unique feature of M.xanthus being predatory towards other bacteria is of great use for studies. This feature has been hypothesized to be utilized in order to predate for other harmful bacteria. Many of the secondary metabolites secreted by M. xanthus are utilized to lyse other soil microbes. One of the many purposes these secondary metabolites serve is the ability to target pathogenic fungi in plants. It is also capable of producing antibiotics which fight molds, yeasts and enterobacteria.[11]
Current research
"Identification of Additional Players in the Alternative Biosynthesis Pathway to Isovaleryl-CoA in the Myxobacterium Myxococcus xanthus Helge B. Bode,[a] Michael W. Ring,[a] Gertrud Schwr, Matthias O. Altmeyer, Carsten Kegler, Ivy R. Jose, Mitchell Singer, and Rolf Miller (2009)”
In this research, there has been found an alternative pathway to Isovaleryl - CoA in M. xanthus. The alternative pathway that has been discovered is 3-hydroxy-3-methylglutaryl-CoA synthase (MvaS), which is persuaded in mutants with non functional leucine degradation or during the formation of fruiting bodies. Leucine is a major fatty acid that M.xanthus requires for growth. . In M.xanthus the biosynthesis of iso-FAs needs isiovaleryl-CoA (IV-CoA). The synthesis could not happen without this enzyme. In this research presents the findings of another pathway of IV-CoA. Experiments having to do with feeding in M.xanthus and stigmalella aurantiaca, led to the alternative pathway findings. The manner in which this theory is tested is by presenting the bacteria with and environment with and without leucine. This confirmed that 3-hydroxy-3-methylglutaryl-CoA synthase (MvaS). This enzyme is also responsible for the formation of HMG-CoA from acetyl CoA.[12]
"Genetic circuitry controlling motility behaviors of Myxococcus xanthus, Ta, m Mignot1 and John R. Kirby2 (2008)"
M.xanthus contains two motility systems type IV-pilli and mucus secreting system, which form focal adhesion sites. Current locomotion studies have revealed that the synchronizing of both motor systems is due to spatial oscillation of motility proteins. M. xanthus predation tactics are being studied with results such as: a sort of cell reversal motion which cause a wave like effect, directing towards the prey. The cells coordinate to form this synchronizing mechanism. During different environment there is a quick response in motility systems. This synchronization is thought to contain at least three chemosensory systems that are very similar to eukaryotic cell signaling proteins. This research its speculated that motility system is directed by oscillations of motility proteins. Type IV-Pilli pulls cell forward, by removing this system, its how recognition of other system was discovered. This is the mucous secreting part of motility, which gives the bacteria a gliding motion. The mechanism of gliding is still not yet discovered. By in vivo testing of pillus associated proteins (Frz-GFP) is how the oscillations were graphed. The synchronization of both systems is supported by the results from Ftz, Aglz and RomR are the main elements in this cycle. This resulted from viewing FrzS, which remained coupled to directional movement where gliding (not TFP) is the main source of motility. When viewing RomR-mDSRed and Frz S-GFP in a cell, each protein built up with comparable kinetics at each pole and reached symmetrical status at the same time. This evidence supports the theory that the motility system of M.xanthus is by synchronizations of this motility system.[13]
"Site-specific receptor methylation of FrzCD in Myxococcus xanthus is controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase, Ansley E. Scott, Eric Simon, Samuel K. Park, Philip Andrews2 and David R. Zusman1 (2008)"
There is also a correlation between the ripple wavelength and amount of prey available. High amounts prey there are shorter wavelengths, lower amounts of prey cause a longer wavelength. Current research has positive results on the identification of the pathway responsible for chemotaxis in this bacteria. The assumed pathway is Frz, which activates FrCD, a chemotaxis protein. Movements of M.xanthus is regulated by Frz chemosensory system. This system controls cell reversals. Frz pathway requires activity of FrzCD. FrzCD is required for swarming and the morph to fruiting bodies, its also methyl-accepting chemotaxis protein. FrzF is a methyl transferase (CheR). Frzt contains and additional domain with three tetra trico-peptide repeats (TPRs). To test the role of TPRs, TPRs were removed in full full length Frz. The results were, the pattern of receptor methylation determined receptor activity. Methylation can activate and inactivate the receptor activity.[14]
References
- ↑ Shi Lab (30 November 2004). Myxococcus xanthus. UCLA Department of Microbiology, Immunology, and Molecular Genetics. Retrieved on 28 November 2013.
- ↑ The social lifestyle of myxobacteria, Arthur L. Koch and David White
- ↑ Tam Mignot1 and John R. Kirby2, (2008), Genetic circuitry controlling motility behaviors of Myxococcus xanthus
- ↑ http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=250
- ↑ http://www.nytimes.com/2008/11/04/science/04obswarm.html?_r=1
- ↑ http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=250
- ↑ http://microbewiki.kenyon.edu/index.php/Mycoccus_xanthus
- ↑ http://www.medicine.uiowa.edu/CCOM/news/video-predataxis
- ↑ http://microbewiki.kenyon.edu/index.php/Mycoccus_xanthus
- ↑ http://mcb.berkeley.edu/labs/zusman/zusman%20lab%20web%20page/Myxococcus%20xanthus.html
- ↑ http://www.bio.indiana.edu/facultyresearch/faculty/Velicer.html
- ↑ Helge B. Bode,[a] Michael W. Ring,[a] Gertrud Schwr, Matthias O. Altmeyer, Carsten Kegler, Ivy R. Jose, Mitchell Singer, and Rolf Miller (2009), dentification of Additional Players in the Alternative Biosynthesis Pathway to Isovaleryl-CoA in the Myxobacterium Myxococcus xanthus
- ↑ Tam Mignot1 and John R. Kirby2, (2008), Genetic circuitry controlling motility behaviors of Myxococcus xanthus
- ↑ Ansley E. Scott, Eric Simon, Samuel K. Park, Philip Andrews2 and David R. Zusman1, (2008) Site-specific receptor methylation of FrzCD in Myxococcus xanthus is controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase